Feature Extraction and Data Preprocessing
- Extraction: Extracting features from “raw” data
- Transformation: Scaling, converting, or modifying features
- Selection: Selecting a subset from a larger set of features
1) Load Data to DataFrame
- Load raw data to DataFrame, for example let's load diabets.csv file into DataFrame:
val inputFile = args(0);
//Initialize SparkSession
val sparkSession = SparkSession
.builder()
.appName("spark-read-csv")
.master("local[*]")
.getOrCreate();
//Read CSV file to DF and define scheme on the fly
val patients = sparkSession.read
.option("header", "true")
.option("delimiter", ",")
.option("nullValue", "")
.option("treatEmptyValuesAsNulls", "true")
.option("inferSchema", "true")
.csv(inputFile)
patients.show(100)
patients.printSchema()
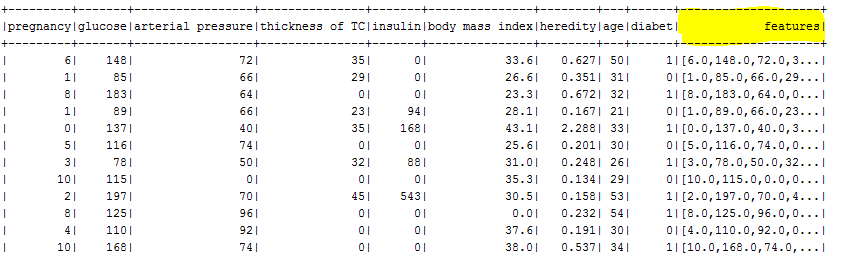
As a result you will see :
2) Extract features and prepare training data
To build a classifier model, you first extract the features that most contribute to the classification:
The features for each item consists of the fields shown below:
Label → diabet: 0 or 1
Features → { "pregnancy", "glucose", "arterial pressure", "thickness of TC", "insulin", "body mass index", "heredity", "age"}
In order for the features to be used by a machine learning algorithm, the features are transformed and put into Feature Vectors, which are vectors of numbers representing the value for each feature.
VectorAssembler
Below a VectorAssembler is used to transform and return a new DataFrame with all of the feature columns in a vector column
//Feature Extraction
val DFAssembler = new VectorAssembler().
setInputCols(Array(
"pregnancy", "glucose", "arterial pressure",
"thickness of TC", "insulin", "body mass index",
"heredity", "age")).
setOutputCol("features")
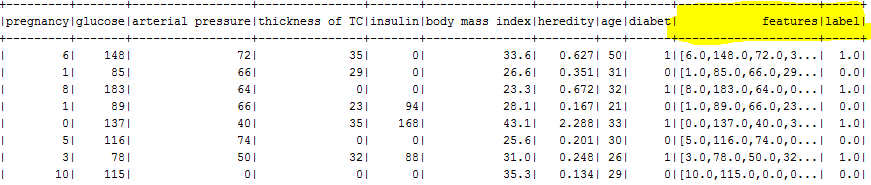
val features = DFAssembler.transform(patients)
features.show(100)
StringIndexer
We also have to use a StringIndexer to return a Dataframe with the clas (malignant or not) column added as a label.
val labeledTransformer = new StringIndexer().setInputCol("diabet").setOutputCol("label")
val labeledFeatures = labeledTransformer.fit(features).transform(features)
labeledFeatures.show(100)
3) Split data into training and testing datasets:
// Split data into training (60%) and test (40%)
val splits = labeledFeatures.randomSplit(Array(0.6, 0.4), seed = 11L)
val trainingData = splits(0)
val testData = splits(1)